Convert point environment data to a raster of majority-environment classes
Source:R/calc_enviro_variables.R
classRast.RdGiven point occurrences of environmental categories, classRast generates
a raster grid with cell values specifying the majority environment therein.
Arguments
- grid
A
SpatRasterto use as a template for the resolution, extent, and coordinate reference system of the returned object. Values can be empty.- dat
Either a
data.frameormatrixfor whichxyandenvare column names, or an empty argument.- xy
A vector specifying the name or numeric position of columns in
datcontaining coordinates, ifdatis supplied, or a 2-columndata.frameormatrixof coordinate values.- env
The name or numeric position of the column in
datcontaining a categorical environmental variable, ifdatis supplied, or a vector of environmental values.- cutoff
The (decimal) proportion of incidences of an environmental category above which a cell will be assigned as that category.
cutoffmust be greater than 0.5.
Details
The cutoff threshold is an inclusive bound: environmental incidence
proportions greater than or equal to the cutoff will assign cell values
to the majority environmental class. For instance, if category A represents
65% of occurrences in a cell and cutoff = 0.65, the returned value for the
cell will be A. If no single category in a cell meets or exceeds the
representation necessary to reach the given cutoff, the value returned
for the cell is indet., indeterminate.
Cells lacking environmental occurrences altogether return NA values.
The env object can contain more than two classes, but in many cases it will
be less likely for any individual class to attain an absolute majority the
more finely divided classes are. For example, if there are three classes,
A, B, and C, with relative proportions of 20%, 31%, and 49%, the cell value
will be returned as indet. because no single class can attain a cutoff
above 50%, despite class C having the largest relative representation.
Missing environment values in the point data should be coded as NA,
not e.g. 'unknown'. classRast() ignores NA occurrences when tallying
environmental occurrences against the cutoff. However, NA occurrences
still count when determining NA status of cells in the raster: a cell
containing occurrences of only NA value is classified as indet., not NA.
That is, any grid cell encompassing original point data is non-NA.
Antell and others (2020) set a cutoff of 0.8, based on the same threshold
Nürnberg and Aberhan (2013) used to classify environmental preferences for taxa.
The coordinates associated with points should be given with respect to the
same coordinate reference system (CRS) of the target raster grid, e.g. both
given in latitude-longitude, Equal Earth projected coordinates, or other CRS.
The CRS of a SpatRaster object can be retrieved with terra::crs()
(with the optional but helpful argument describe = TRUE).
Examples
library(terra)
# work in Equal Earth projected coordinates
prj <- 'EPSG:8857'
# generate point occurrences in a small area of Northern Africa
n <- 100
set.seed(5)
x <- runif(n, 0, 30)
y <- runif(n, 10, 30)
# generate an environmental variable with a latitudinal gradient
# more habitat type 0 (e.g. rock) near equator, more 1 (e.g. grassland) to north
env <- rbinom(n, 1, prob = (y-10)/20)
env[env == 0] <- 'rock'
env[env == 1] <- 'grass'
# units for Equal Earth are meters, so if we consider x and y as given in km,
x <- x * 1000
y <- y * 1000
ptsDf <- data.frame(x, y, env)
# raster for study area at 5-km resolution
r <- rast(resolution = 5*1000, crs = prj,
xmin = 0, xmax = 30000, ymin = 10000, ymax = 30000)
binRast <- classRast(grid = r, dat = ptsDf, xy = c('x', 'y'),
env = 'env', cutoff = 0.6)
binRast
#> class : SpatRaster
#> dimensions : 4, 6, 1 (nrow, ncol, nlyr)
#> resolution : 5000, 5000 (x, y)
#> extent : 0, 30000, 10000, 30000 (xmin, xmax, ymin, ymax)
#> coord. ref. : WGS 84 / Equal Earth Greenwich (EPSG:8857)
#> source(s) : memory
#> categories : mainClass
#> name : mainClass
#> min value : grass
#> max value : indet.
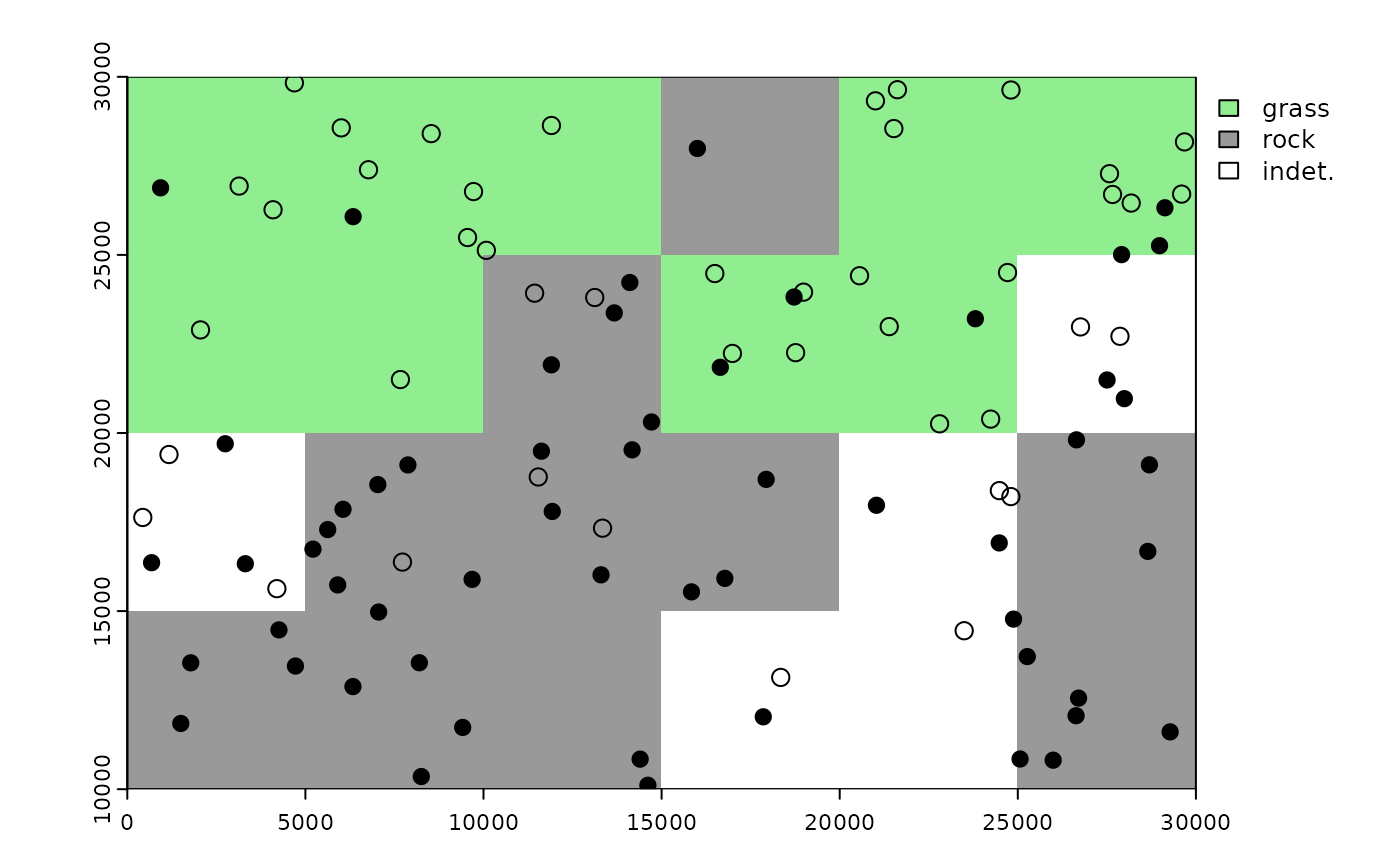
# plot environment classification vs. original points
plot(binRast, col = c('lightgreen', 'grey60', 'white'))
points(ptsDf[env=='rock', ], pch = 16, cex = 1.2) # occurrences of given habitat
points(ptsDf[env=='grass',], pch = 1, cex = 1.2)
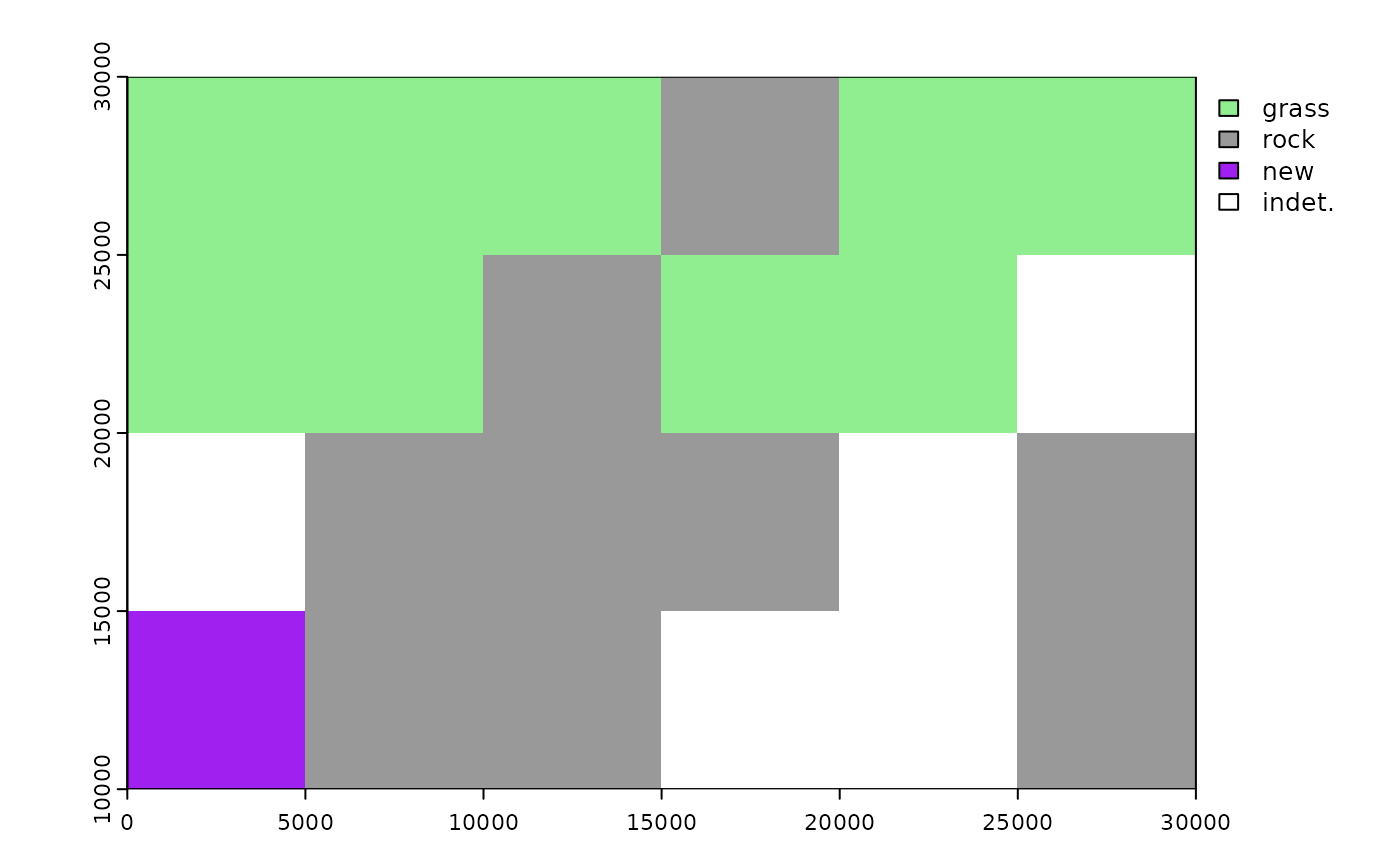
 # classRast can also accept more than 2 environmental classes:
# add a 3rd environmental class with maximum occurrence in bottom-left grid cell
newEnv <- data.frame('x' = rep(0, 10),
'y' = rep(10000, 10),
'env' = rep('new', 10))
ptsDf <- rbind(ptsDf, newEnv)
binRast <- classRast(grid = r, dat = ptsDf, xy = c('x', 'y'),
env = 'env', cutoff = 0.6)
plot(binRast, col = c('lightgreen', 'grey60', 'purple', 'white'))
# classRast can also accept more than 2 environmental classes:
# add a 3rd environmental class with maximum occurrence in bottom-left grid cell
newEnv <- data.frame('x' = rep(0, 10),
'y' = rep(10000, 10),
'env' = rep('new', 10))
ptsDf <- rbind(ptsDf, newEnv)
binRast <- classRast(grid = r, dat = ptsDf, xy = c('x', 'y'),
env = 'env', cutoff = 0.6)
plot(binRast, col = c('lightgreen', 'grey60', 'purple', 'white'))